Annotated draft genome assemblies of the nine bacterial isolates from?
Nusrat Sultana (Ph.D.) Associate Professor Department of Botany Jagannath University
Annotated draft genome assemblies of the nine bacterial isolates from?
Objectives of the research work
1. To characterize the bacterial communities colonizing the rhizosphere of Panchabrihi rice roots.
2. To evaluate the plant growth–promoting potential of the three isolated bacterial strains.
3. To identify and analyze the antibiotic resistance genes associated with these bacterial isolates.
Annotated draft genome assemblies of the nine bacterial isolates from?
Bioinformatics Pipeline used for this research work
1. FastQC on
k1-AC_S15_R1_001.fastq k1-AC_S15_R2_001.fastq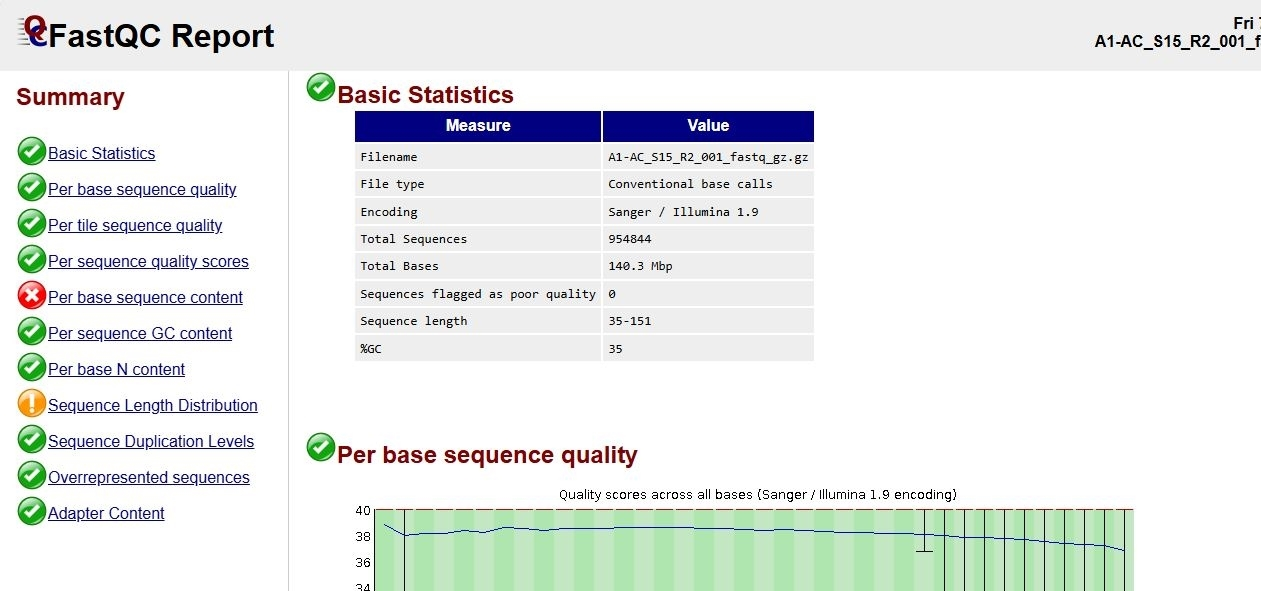
Annotated draft genome assemblies of the nine bacterial isolates from?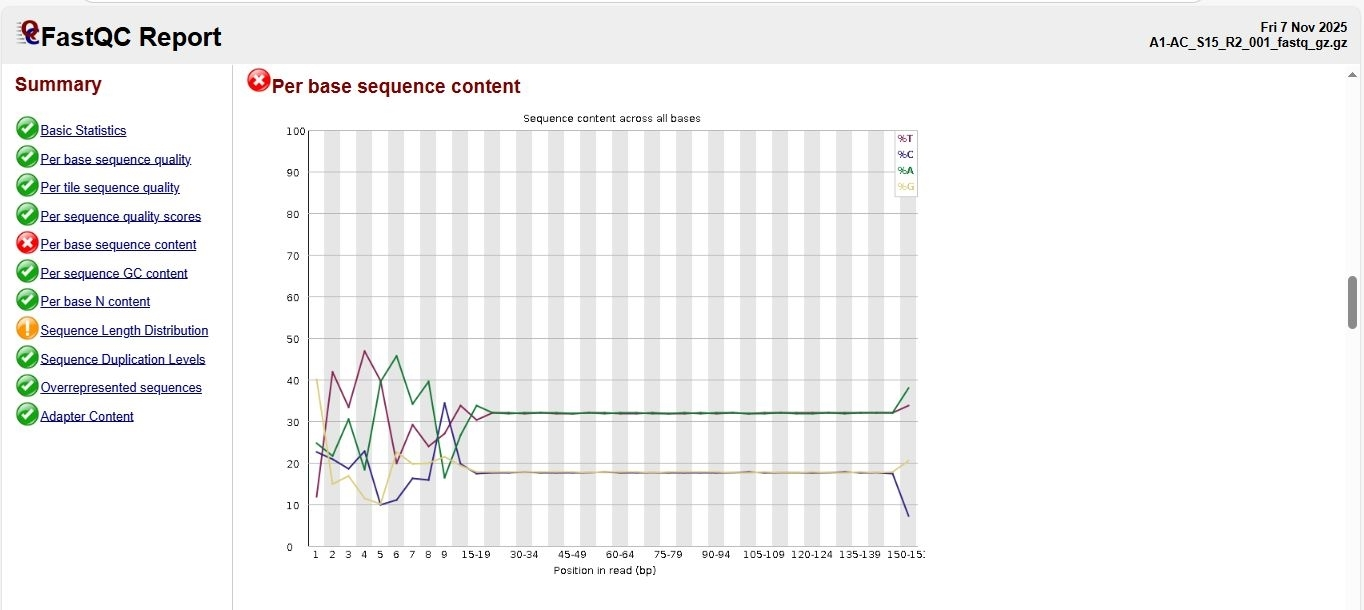
Annotated draft genome assemblies of the nine bacterial isolates from?
2. fastq_paired_end_joiner
k1-AC_S15_R1_001.fastq k1-AC_S15_R2_001.fastq
<br>
k1 (112 Mb)
3. Fastp on k1
Condition: adapter_trimming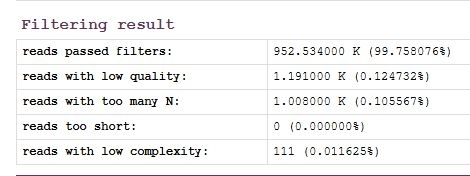
Enable overrepresented analysis quality_filtering_options
Enable low complexity filter Global trimming option 1-20
Annotated draft genome assemblies of the nine bacterial isolates from?
4. Assembly with MEGAHIT Condition: Minimum length of contigs to output 1500 bp
5. QUAST on Assembly Condition; Assembly of the metagenome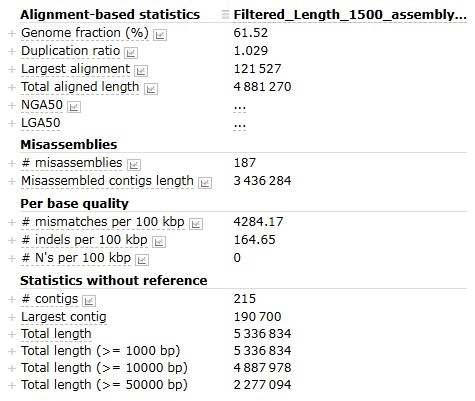
Annotated draft genome assemblies of the nine bacterial isolates from?
6. MetaBAT2, an automated metagenome binning software tool were used to reconstruct single genomes from microbial communities for subsequent analyses of uncultivated microbial species.
7. CheckM v1 estimates genome completeness and contamination on every single genome based on the presence or absence of marker genes, i.e., genes that are typically ubiquitous and single copy.
<br>
Default parameter
Default parameter
Annotated draft genome assemblies of the nine bacterial isolates from?
8. QUAST on Assembly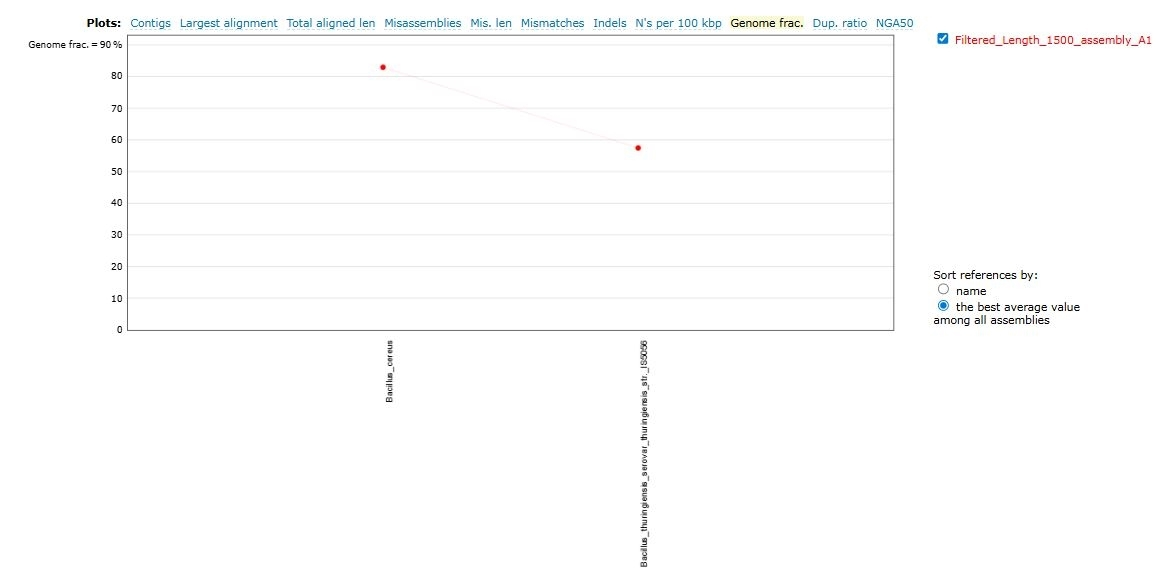
Annotated draft genome assemblies of the nine bacterial isolates from?
9. BUSCO on assembly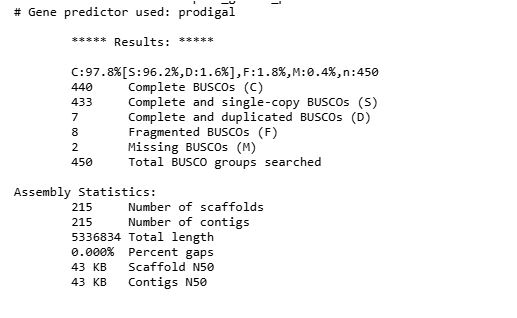
Annotated draft genome assemblies of the nine bacterial isolates from?
10. Bakta: Assembly annotation on genomes using Bakta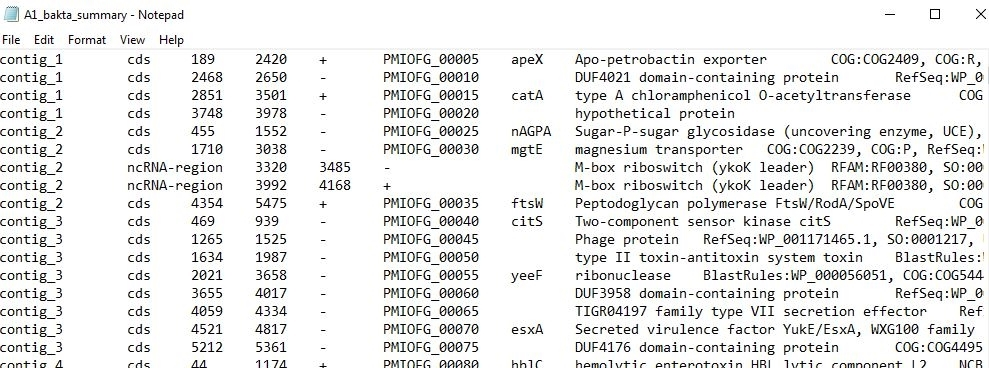
Annotated draft genome assemblies of the nine bacterial isolates from?
K1-2: Acinetobacter schindleri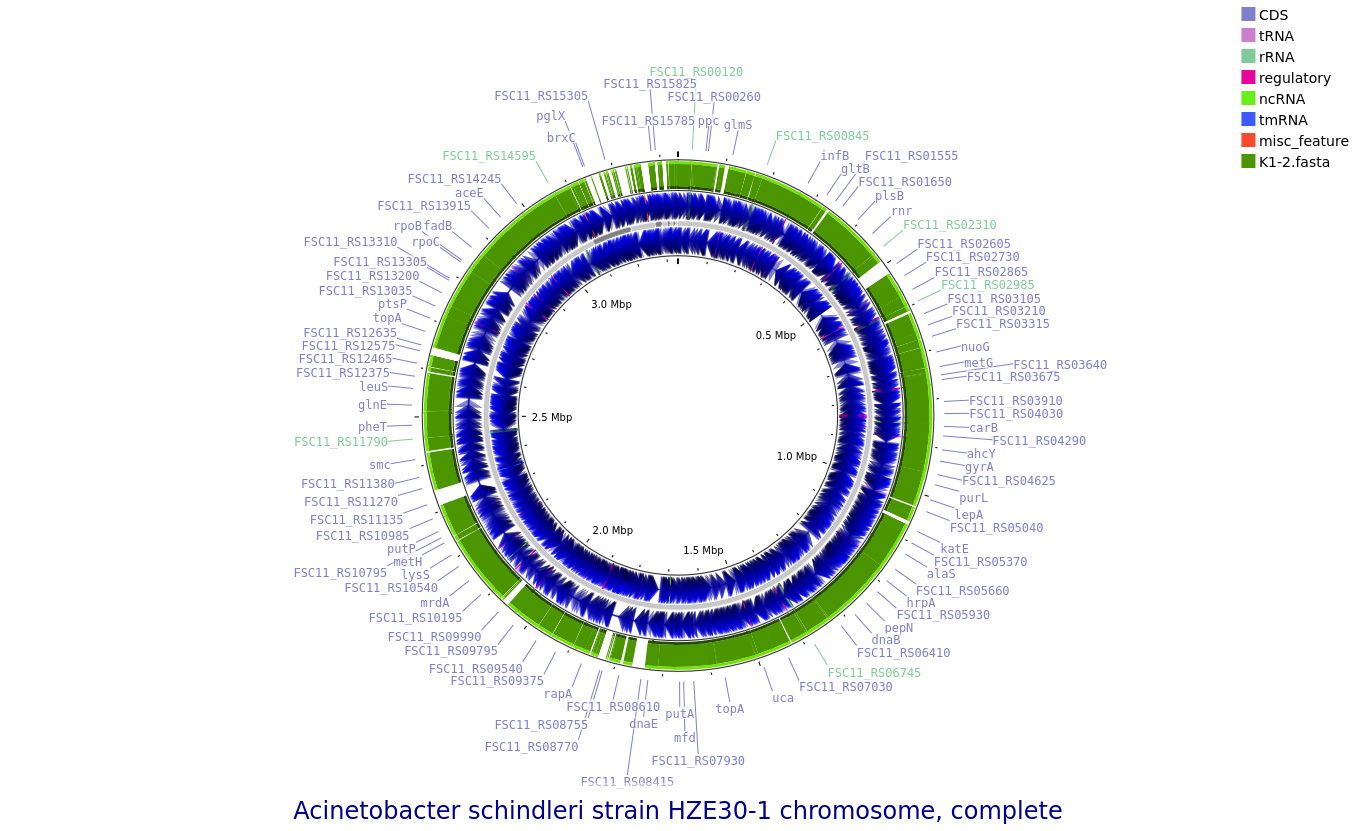
Annotated draft genome assemblies of the nine bacterial isolates from?
K2: Pseudomonas aeruginosa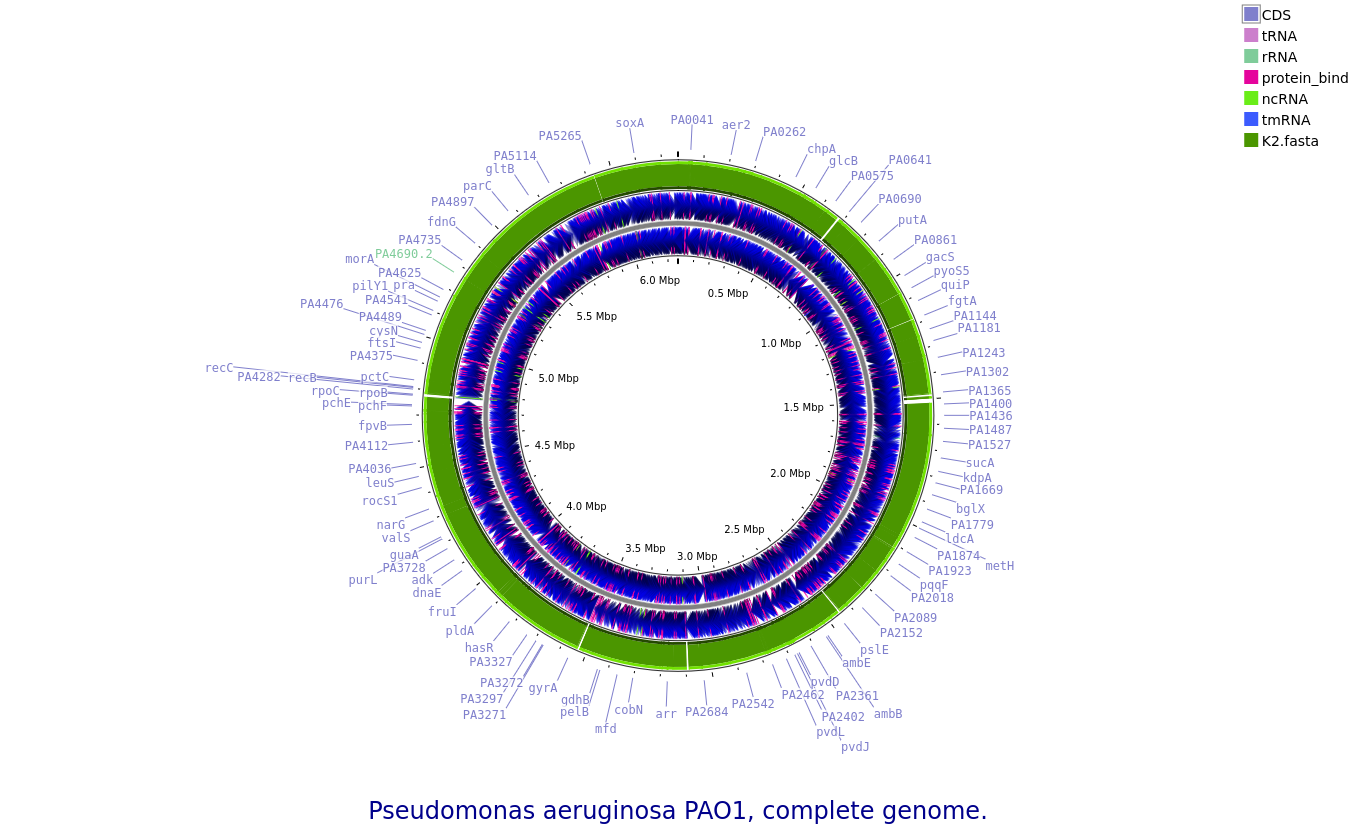
Annotated draft genome assemblies of the nine bacterial isolates from?
KK3-1: Mangrovibacter phragmitis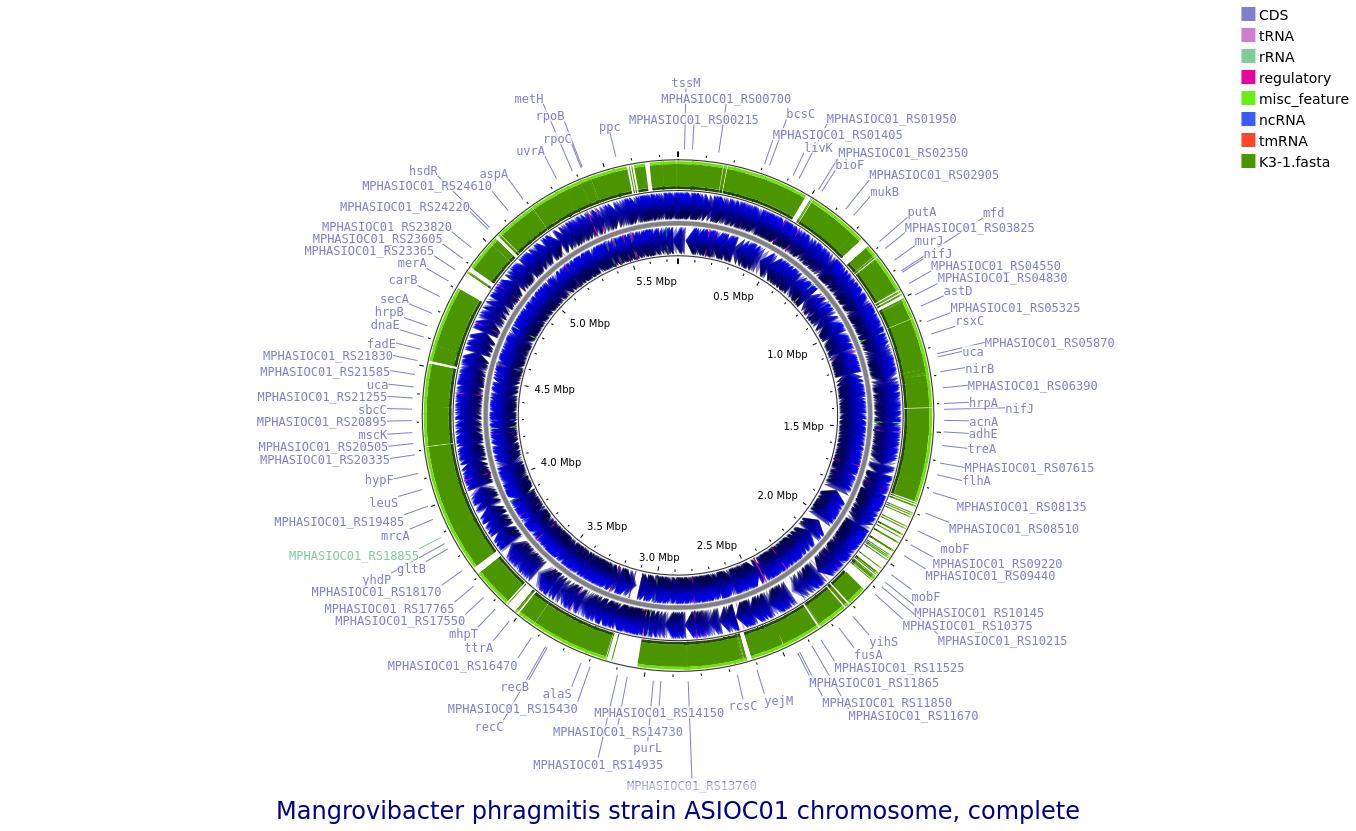
Annotated draft genome assemblies of the nine bacterial isolates from?
KK3-2: Exiguobacterium mexicanum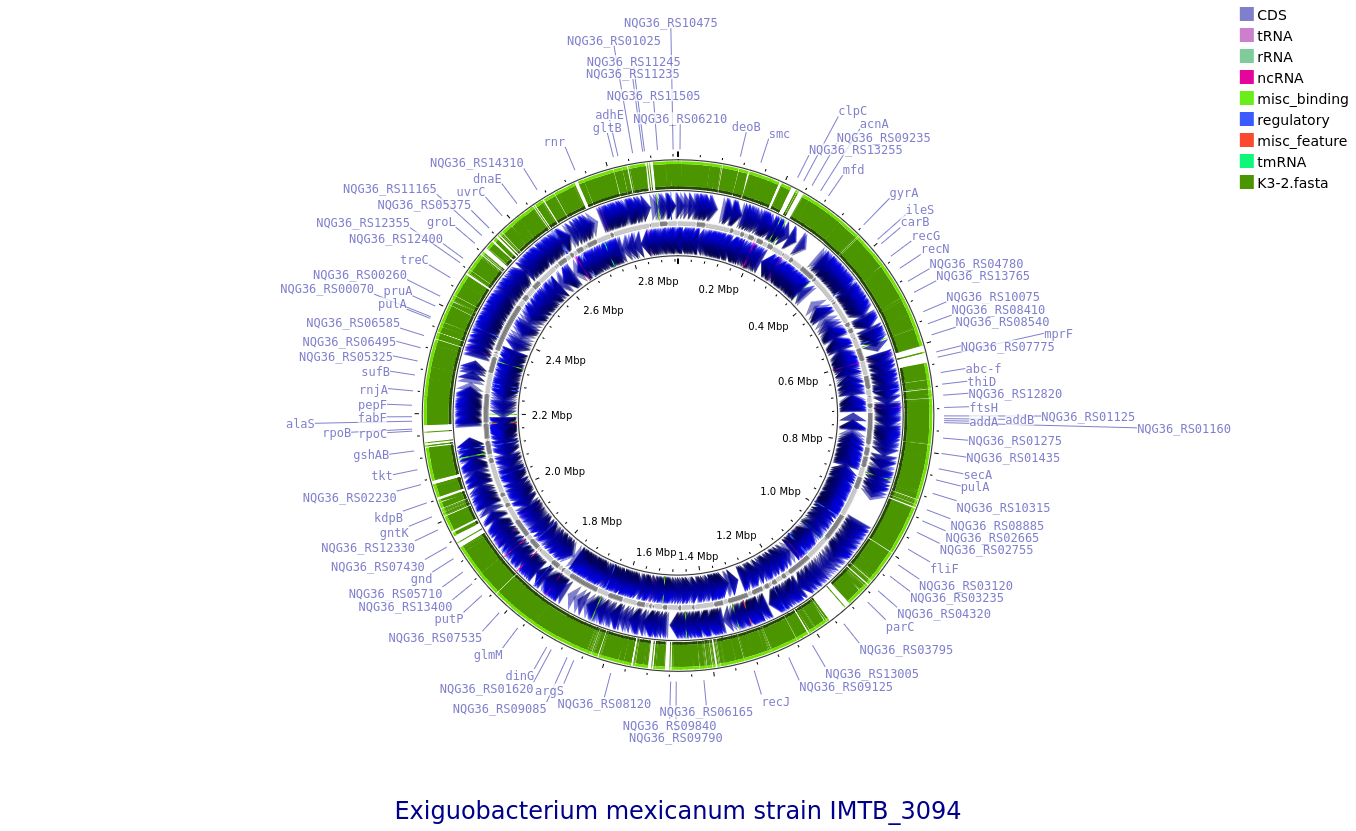
Annotated draft genome assemblies of the nine bacterial isolates from?
K4-1: Bacillus cereus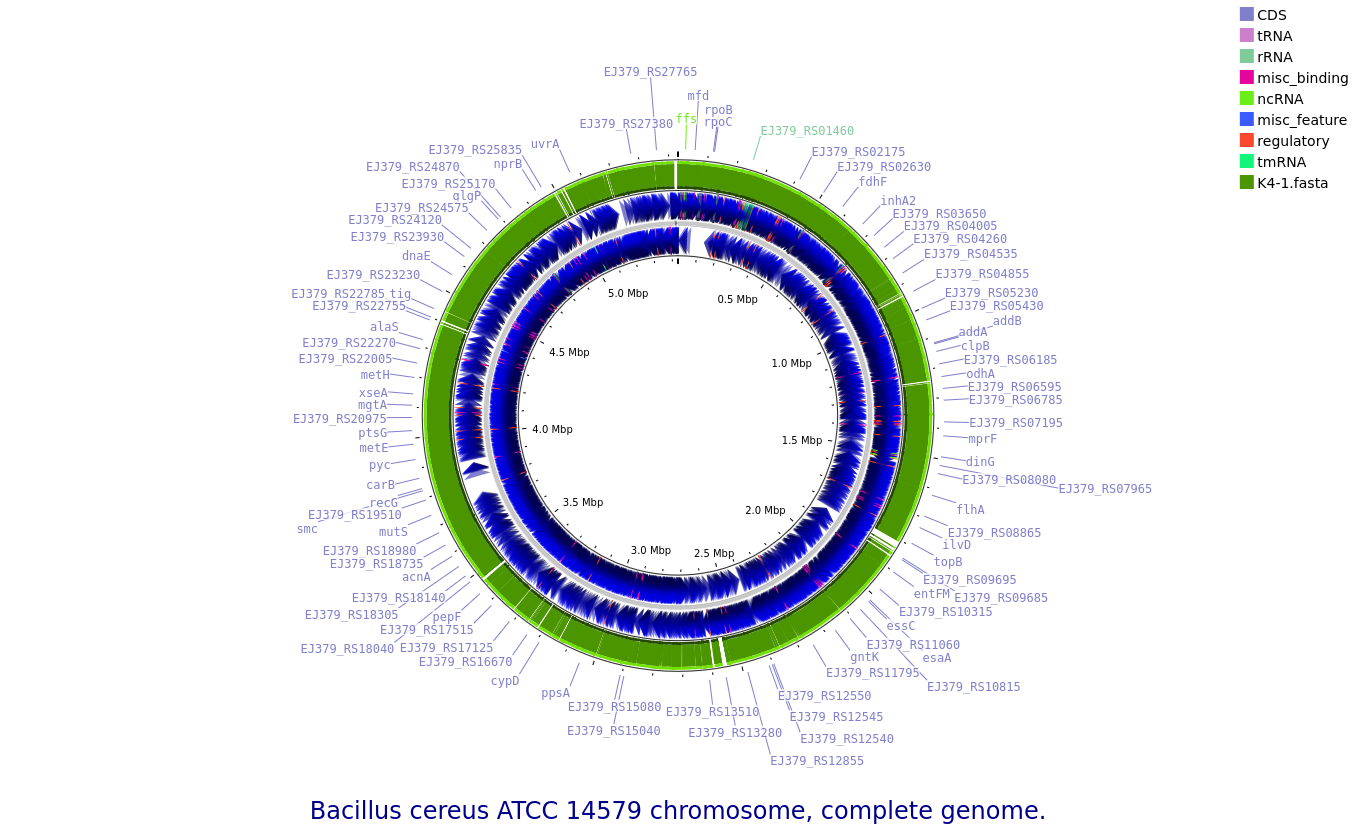
Annotated draft genome assemblies of the nine bacterial isolates from?
K6-1: Bacillus safensis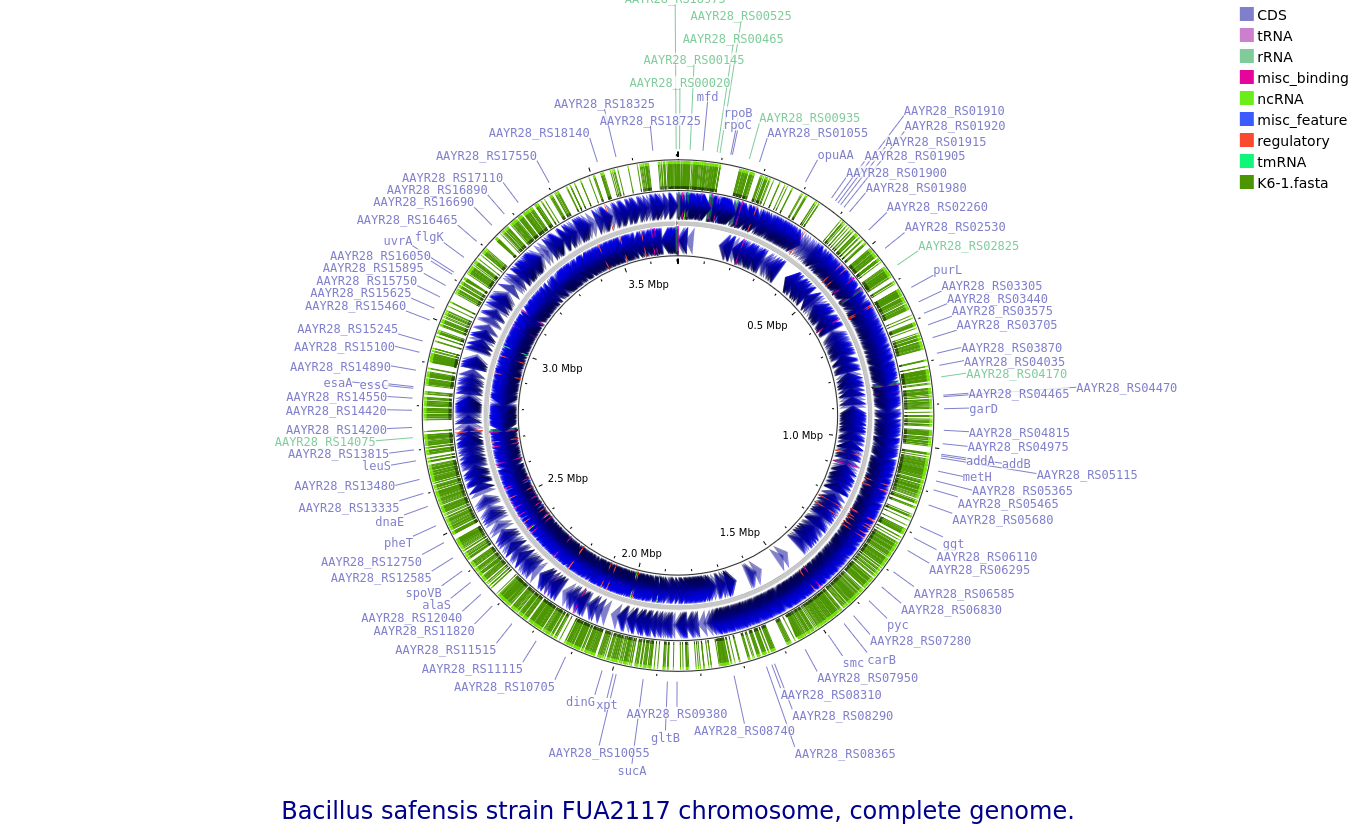
Annotated draft genome assemblies of the nine bacterial isolates from?
K7-2: Bacillus enclensis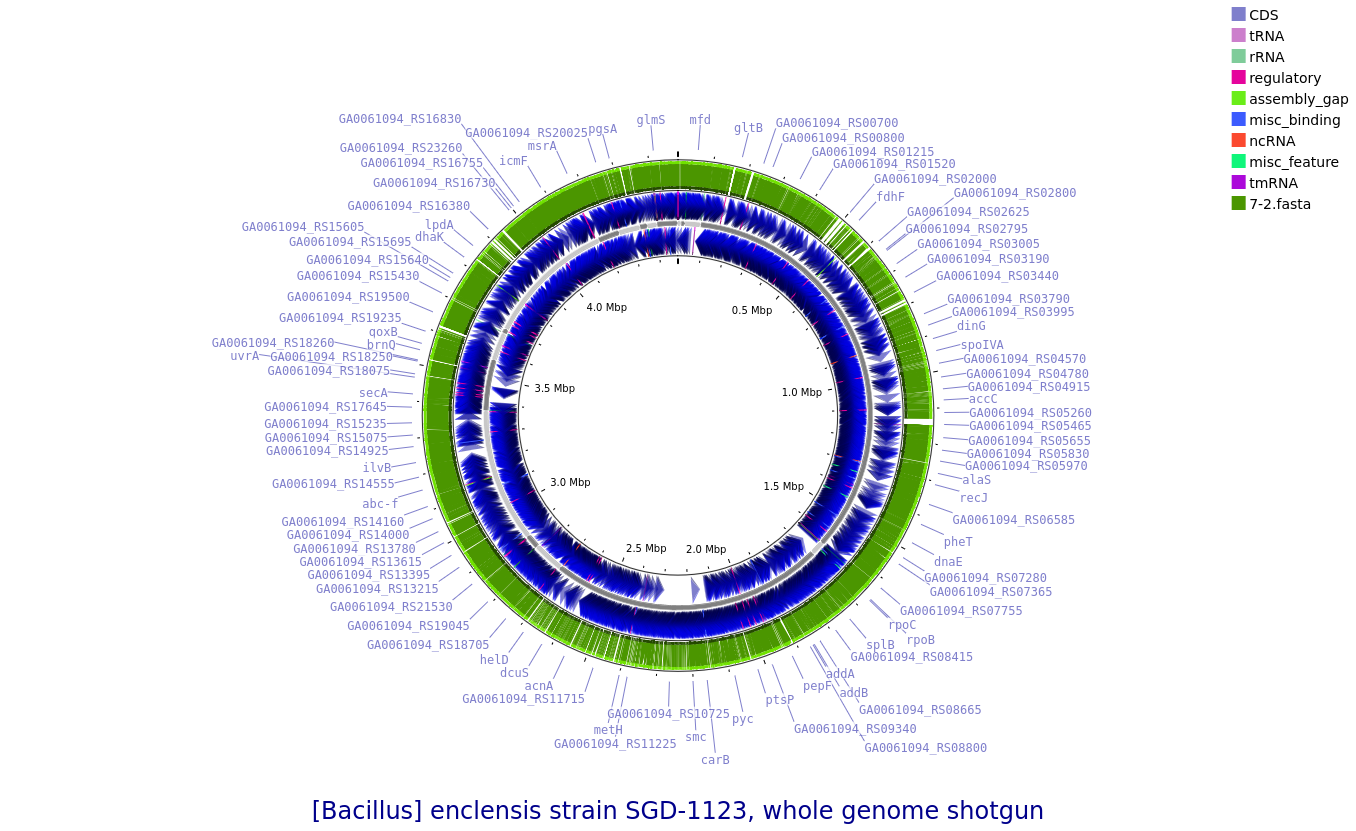
Annotated draft genome assemblies of the nine bacterial isolates from?
K8: Bacillus velezensis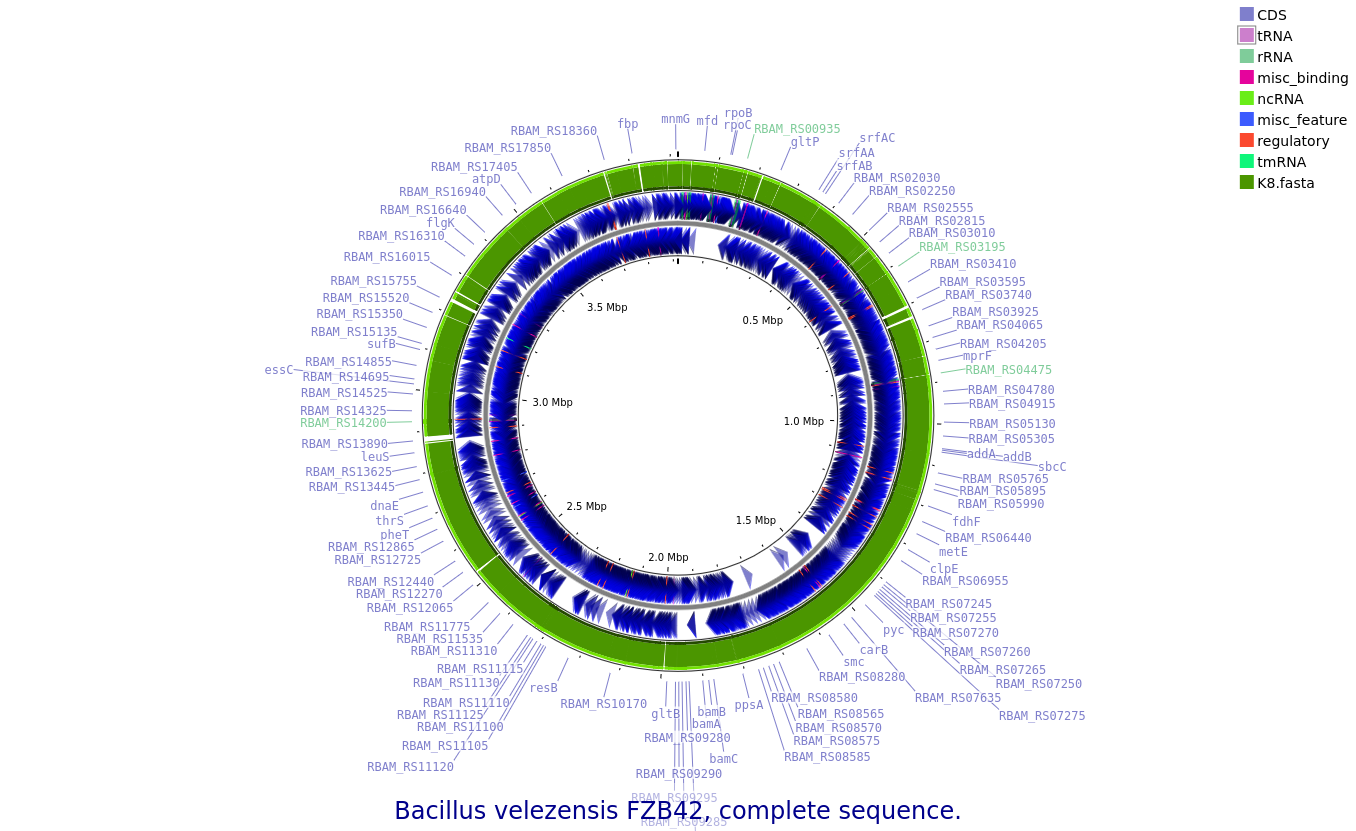
Annotated draft genome assemblies of the nine bacterial isolates from?
K10-2: Cytobacillus firmus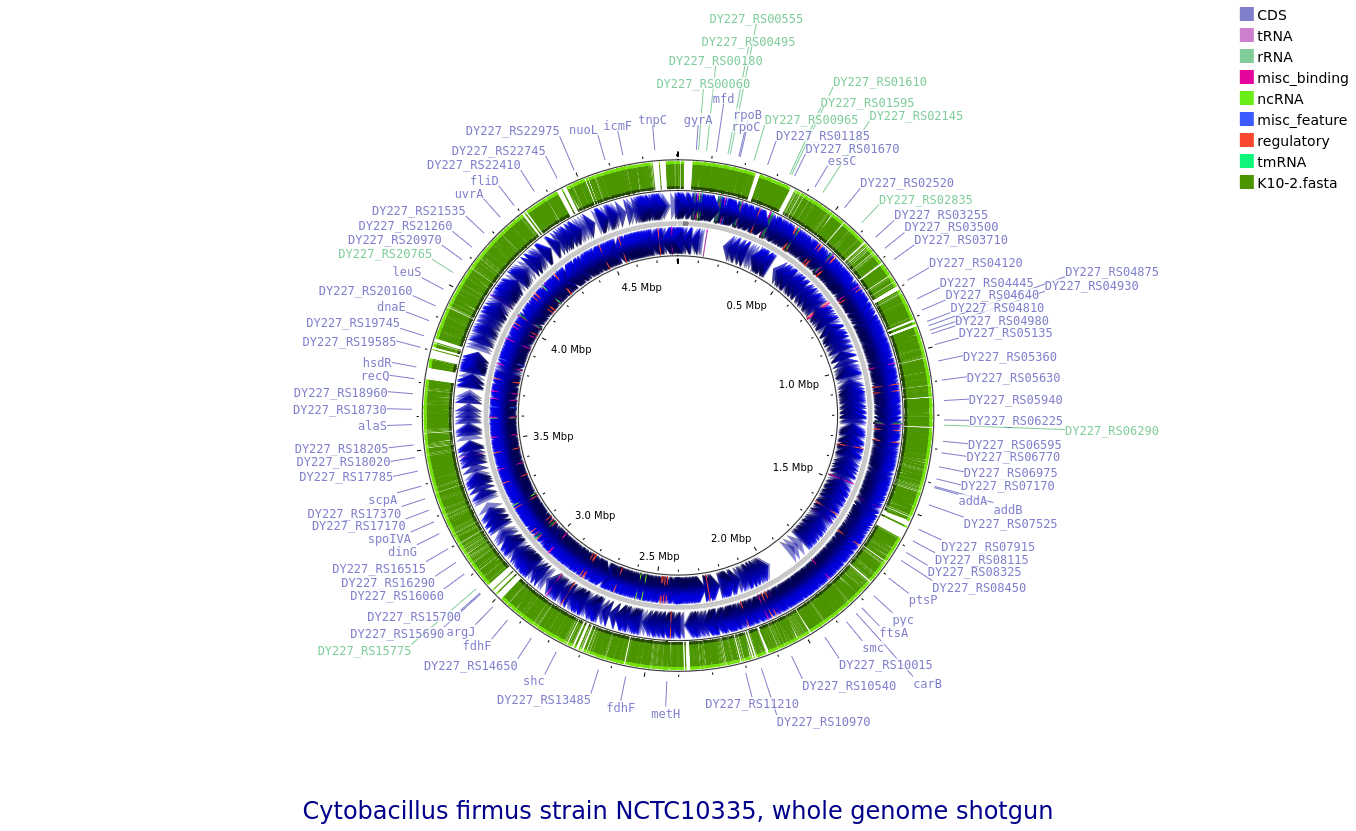
Annotated draft genome assemblies of the nine bacterial isolates from?
Table 1: Genome characteristics of nine bacterial samples (K1-K8 and K10)
SL No Sampl Organism e
<br>
CheckM Genome completeness
<br>
Complete Busco (%)
<br>
Number of contigs
<br>
Total Contig length N50 (KB)
(MB)
(%)
1 K1-2 Acinetobacter schindleri 100
2 K2 Pseudomonas aeruginosa 83.33
3 K3-1 Mangrovibacter phragmitis 100
4 K3-2 Exiguobacterium 54 mexicanum
5 K4-1 Bacillus cereus 100 6 K6-1 Bacillus safensis 100 7 K7-2 Bacillus enclensis 100 8 K8 Bacillus velezensis 100
9 K10-2 Cytobacillus firmus 100
<br>99.4 66 3.1 80
98.2 38 6.2 426
99.8 52 4.9 164
82.9 21 2.5 226
99.3 51 5.5 355 99.6 25 3.7 238 90 535 3.9 12 99.3 15 3.9 580
91.8 701 4.5 8
Annotated draft genome assemblies of the nine bacterial isolates from?
Table 2: Genome characteristics of nine bacterial samples (K1-K8 and K10)
SL Sample Organism No
<br>
Annotate the genome sequence with Prokka
CDS tmRNA tRNA rRNA
1 K1-2 Acinetobacter schindleri 3070 1 65 5 2 K2 Pseudomonas aeruginosa 3070 1 65 5
3 K3-1 Mangrovibacter phragmitis 4505 1 71 4 4 K3-2 Exiguobacterium mexicanum 2650 1 36 2 5 K4-1 Bacillus cereus 5556 1 50 9 6 K6-1 Bacillus safensis 4362 0 59 5 7 K7-2 Bacillus enclensis 3845 0 49 4 8 K8 Bacillus velezensis 3800 1 55 5
9 K10-2 Cytobacillus firmus 4354 1 44 7



